
Multilabel Approach Improves PET/CT Lesion Misidentification!
3 main points
✔️ Demonstrate the effectiveness of a multi-label model
✔️ Improve PET/CT lesion misidentification
✔️ Demonstrate the importance of incorporating multiple labels as well as additional data
Improving Lesion Segmentation in FDG-18 Whole-Body PET/CT scans using Multilabel approach: AutoPET II challenge
written by Gowtham Krishnan Murugesan, Diana McCrumb, Eric Brunner, Jithendra Kumar, Rahul Soni, Vasily Grigorash, Stephen Moore, Jeff Van Oss
(Submitted on 2 Nov 2023)
Comments: AutoPET II challenge paper
Subjects: Image and Video Processing (eess.IV); Computer Vision and Pattern Recognition (cs.CV); Machine Learning (cs.LG)
code:
The images used in this article are from the paper, the introductory slides, or were created based on them.
Introduction
Automated lesion segmentation in FDG-18 whole-body (WB) PET/CT scans using deep learning models can help determine treatment response, optimize dosimetry, and advance the application of treatments in oncology. However, the presence of organs with increased uptake of radiation tracers (chemicals in which one or more atoms have been replaced by a radionuclide), such as the liver, spleen, brain, and bladder, poses a challenge because these regions can be misidentified as lesions by the deep learning model.
To address this issue, this paper proposes a new approach to segment both organs and lesions with the aim of improving the performance of automatic lesion segmentation methods.
Current Status and Issues of PET/CT
Positron emission tomography/computed tomography (PET/CT) is an indispensable tool for tumor imaging, helping to detect metastatic disease at an early stage, quantify metabolic hypertrophic tumors, and contribute significantly to cancer diagnosis, staging, treatment planning and recurrence monitoring.
At the same time, advances in deep learning algorithms have revolutionized the field of medical imaging, facilitating more accurate and efficient segmentation of cancerous lesions in acquired images. These algorithms have the remarkable ability to autonomously depict tumor boundaries even in situations where traditional methodologies fail to segment due to the complexity, variability, and subtlety of the lesion.
However, the uptake of radioactive tracers varies among patients, and the inherent nature of radioactive tracers can lead to high accumulation in normal, metabolically active organs such as the brain, and in purifying organs such as the liver, kidneys, and bladder. Therefore, it is difficult for automated lesion segmentation algorithms to distinguish between normal uptake of radioactive tracers in these organs and true lesions, which is a major challenge.
Hypothesis and Verification
In this paper, wehypothesize that to solve the problem of automatic lesion segmentation algorithms,organs with high uptake ofradioactive tracers can be segmentedalong the lesion so that the model can incorporate the discriminative power of whether the organ is a lesion or not, and we have tested several We are testing this hypothesis in several ways.
Method
Data and Preprocessing
The data used in this paper are whole-body FDG-PET/CT data from 900 patients, including 1,014 studies provided by AutoPET challenge II 2023. In addition, a retained data set consisting of 150 studies (100 of which were from the same hospital as the training database and 50 from another hospital with similar acquisition protocols ) was used as a test data set to evaluate the robustness and generalizability of the algorithm proposed in this paper. from another hospital with a similar acquisition protocol) is used as the test dataset for generalizability assessment.
As a preprocessing step, the CT data is resampled and normalized to PET resolution. In addition, two experts labeled the training and test data, respectively: a radiologist with 10 years of experience in hybrid imaging ( a medical imaging technique that combines two or more different imaging modalities to provide a more comprehensive picture of the body's internal structure and function ) and a radiologist with five years of experience in machine learning research at the University Hospital Tübingen University Hospital, and a radiologist with five years of experience in hybrid imaging and machine learning research at LMU University Hospital in Munich labeled all data.
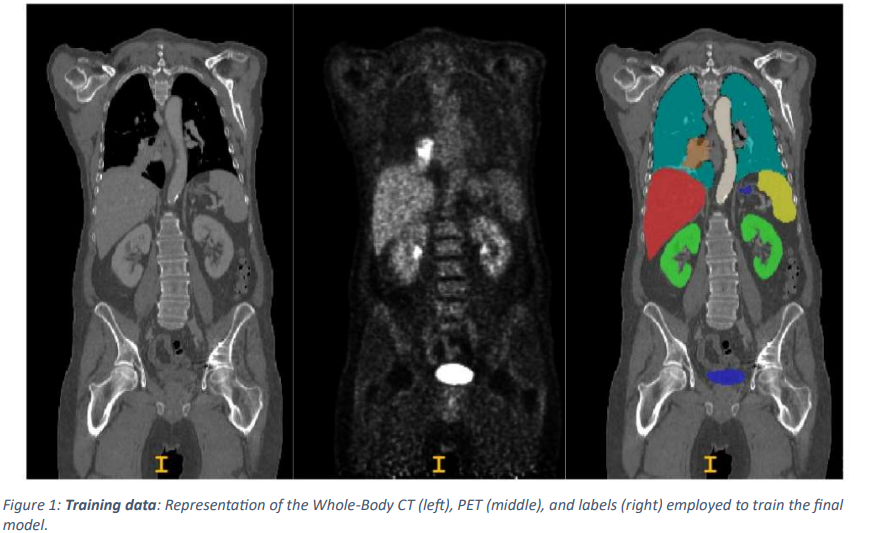
In this paper, the training data is randomly divided into 5 segments and the 3D UNet model is trained within the nnUNet framework to segment multiple organs and lesions.
Impact of adding multiple labels and data
To evaluate the proposed method, we first split thetraining data intotwo groups of experiments(one containing 819 studies and the other containing 195 studies) and then conducted a study to evaluate the effect of adding more labels (liver, spleen, kidney, bladder) and more data on the model performance in segmenting lesions. The studies are being conducted to evaluate the effect of
This paper specifically focuses on two different 3D UNET models designed for lesion segmentation and implements a thorough model training and evaluation process utilizing a 5-segment cross-validation method.
One model is created to isolate lesions (single-label) and the other to segment lesions in combination with other high-intake organs (multi-label). However, the labels for the high intake organs are derived using the publicly available totalsegmentator. We first train both models on a dataset consisting of 819 studies and rigorously evaluate their performance on another dataset consisting of 195 different medical studies. This evaluation allows us to assess the effectiveness of the models in real-world situations.
In addition, to investigate the effect of dataset size on model performance, a subset of 100 studies from the original 819 studies was randomly selected to train two separate models with identical segmentation objectives (segmentation of lesions only: single-label model; segmentation of lesions and high-intake organs: multi-label model). We then evaluate these models using the same 195 study retention dataset to allow us to understand how the dataset size affects the segmentation function.
AutoPET II Challenge
The AutoPET II Challengeuses all of thetrainingdata from1,014 studies to train the multi-label model. Eight additional organs (liver, kidney, bladder, spleen, lung, brain, heart, and stomach) were derived and added to the training data set using the publicly available totalsegmentator package.
Model Architecture
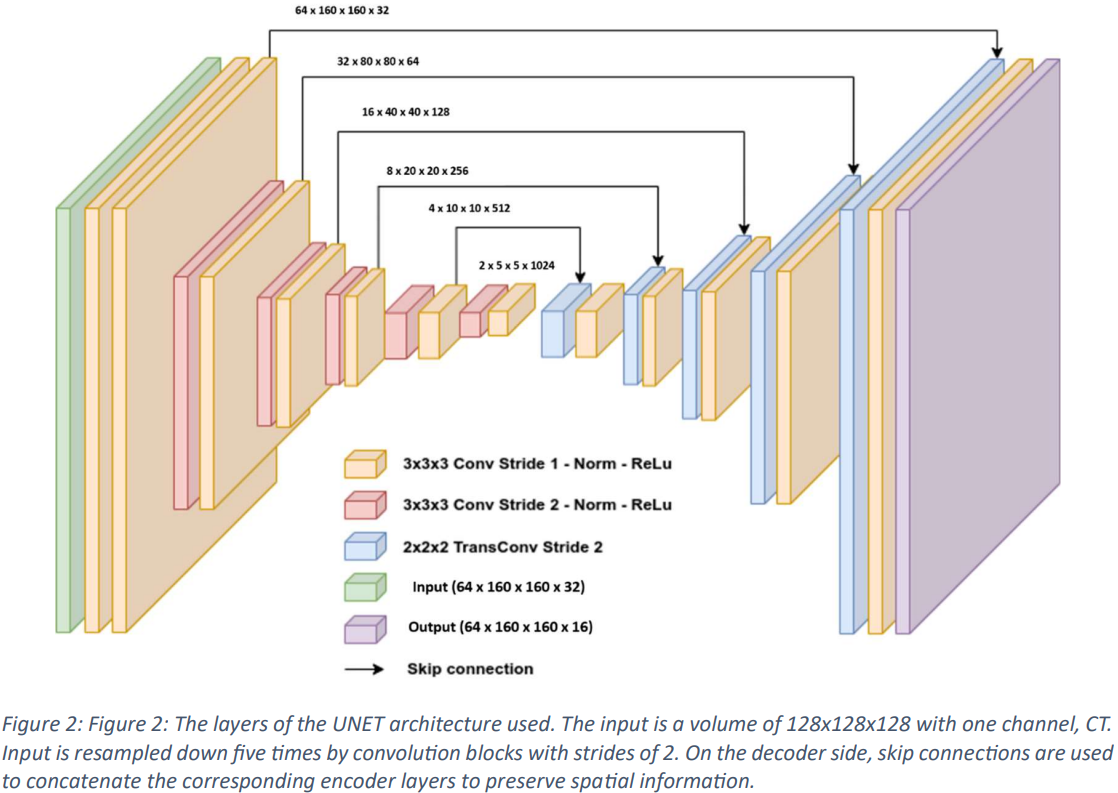
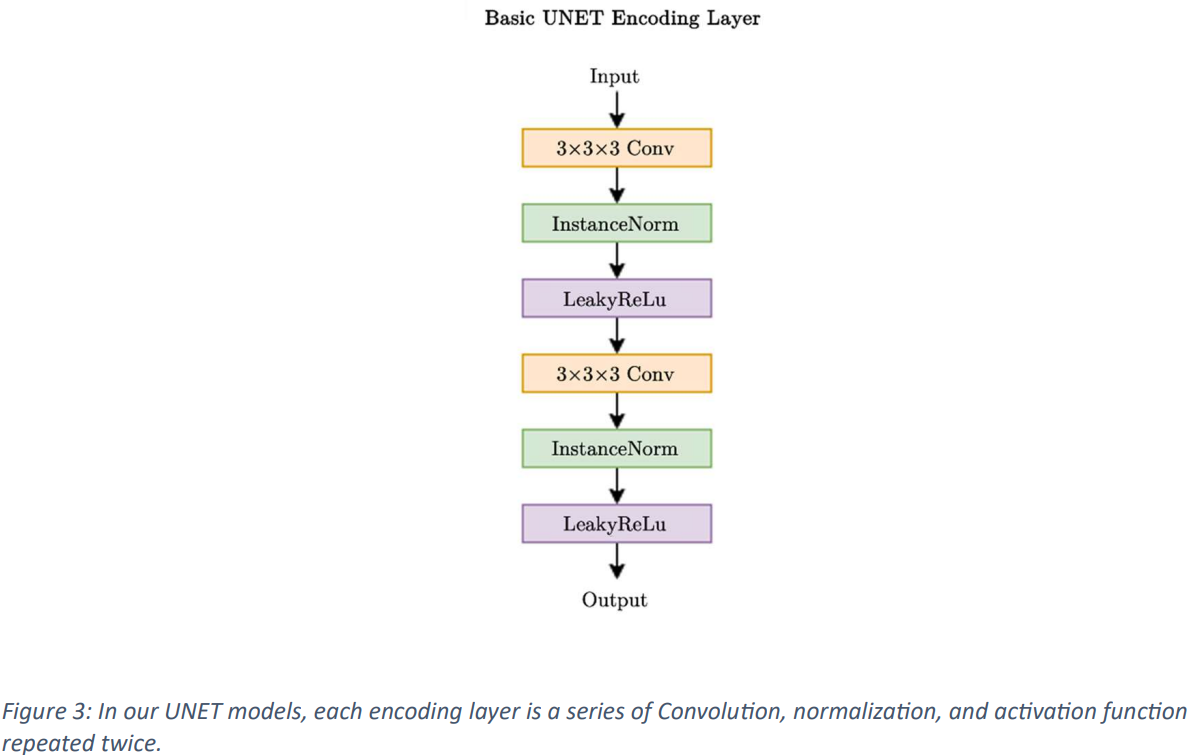
In this paper, a standard variant 3D UNet model is used, incorporating skip connections for training, as shown in Figs. 2 and 3. The input image dimensions are set to 128x128x128 on a single channel, and a CT scan is used as input data. The input is progressively downsampled by 2x stride convolutional blocks through 5 stages, and on the decoder side, skip connections are used to combine the corresponding encoder layers and store spatial information. The network layer incorporates InstanceNorm and utilizes LeakyReLu.
The initial architecture employs 32 feature maps, doubling during each downsampling operation at the encoder and halving again during the inverted convolution at the decoder before reaching a maximum of 1024 feature maps. The decoder output maintains the same spatial dimension as the input and then undergoes a 1x1x1 convolution to produce a single-channel output, which is processed by the SoftMax function. During training, the model is trained over a factor of 5 using a loss function that combines the Duce Sorensen Coefficient (DSC) and weighted cross-entropy error to avoid over-training.
To increase the robustness of the models, extension techniques such as random rotation, random scaling, random elastic deformation, gamma correction, mirroring, and elastic deformation are employed. Each of the five models is trained over 1,000 epochs with a batch size of 8 using the SGD optimizer with a learning rate of 0.01. Performance evaluation includes metrics such as Dice Similarity Coefficient (DSC) and Normalized Surface Dice (NSD) to assess various aspects of the segmentation method.
Verification Results
Impact of adding multiple labels and data
Dice Similarity Coefficient (DSC), False Positive Volumes (FPV), and False Negative Volumes (FNV) are used as evaluation methods in this paper.
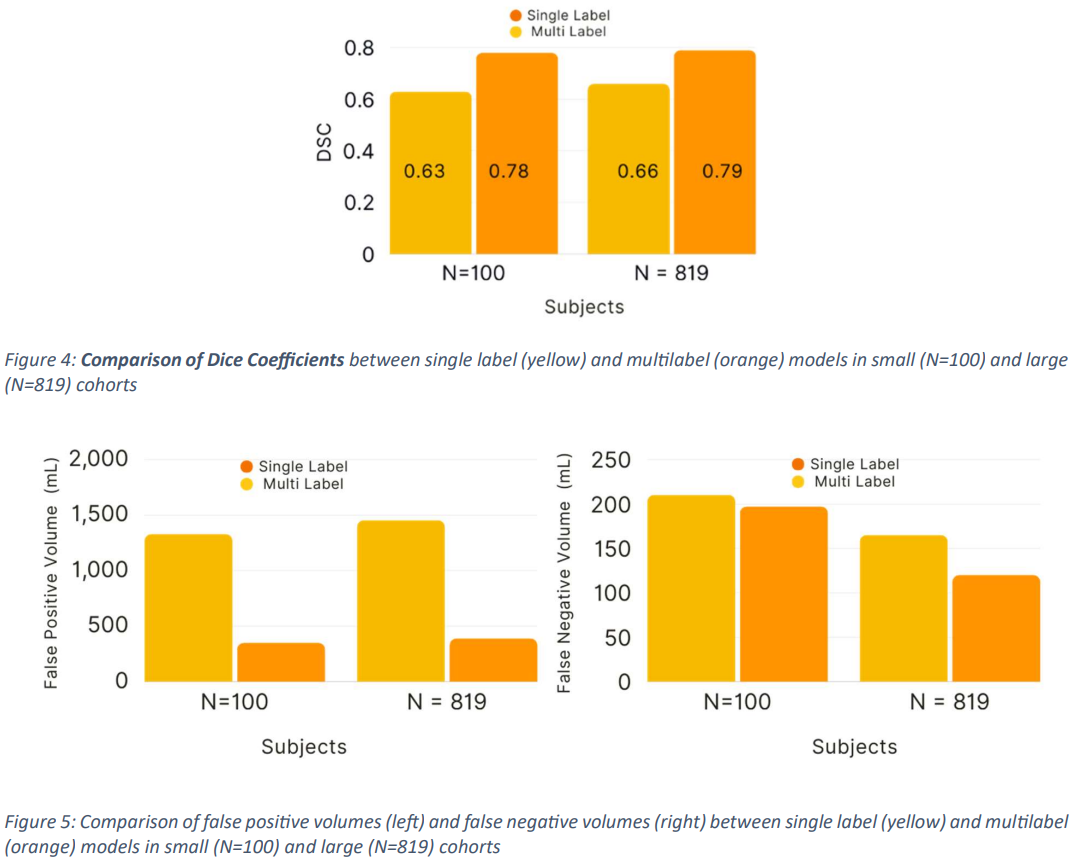
The model trained using a subset of 100 studieshadDSC 0.63+/-0.04 (95% confidence interval (CI): 0.57, 0.69), FPV 1330.45 (95% CI: 165.08, 256.02) and FNV 210.55 (95 % CI: 98.35, 141.88), while the multi-label segmentation achieved DSC 0.78+/-0.02 (95% CI: 0.7, 0.80), FPV 351.13 (161.40, 233.49) and FNV 197.44 (95% CI: 98.35, 141.88).
The model trained using the entire dataset from Study 819 also produced higher DSC scores, specificallyDSC 0.66+/-0.08 (95% CI: 0.54, 0.77), FPV 1456.72 (95% CI: 262.03, 279. 87) for single-label segmentation, DSC 0.79+/-0.02 (95% CI: 98.37, 141.88), and FNV 165.30 (95% CI: 98.37, 141.88) for multilabel segmentation. 87), and FNV 165.30 (95% CI: 98.37, 141.88), for multi-label segmentation, DSC 0.79+/-0.02 (95% CI: 0.77, 0.82), FPV 389.28 (95% CI: 295.83, 482.72), and FNV 120 . 12 (95% CI: 98.35, 141.88 ) (Fig. 4 and Fig. 5) (Tab. 1 ).
From the above, it can be seen that the multi-label segmentation proposed in this paperoutperforms theaccuracy of conventional methods.
AutoPET II Challenge
The validation results from the proposed method in this paper achieved the top ranking in the AutoPET II Challenge by achieving xx dice, yy FPV, and zz FNV.
Summary
In this paper, we present the training and evaluation of two different deep learning models for the purpose of segmenting lesions in medical image data: a single-label model that is designed to focus only on lesion segmentation, and a multi-label model that is designed to segment lesions plus other anatomical structures . The results show the clear effectiveness of the multi-label model.
It also demonstrates the importance of incorporating multiple labels as well as additional data in enhancing the lesion segmentation capabilities of deep learning models.
Because of its effectiveness as well as its versatility, the multilabel approach is expected to develop further in the future.
Categories related to this article



![DrHouse] Diagnostic](https://aisholar.s3.ap-northeast-1.amazonaws.com/media/February2025/drhouse-520x300.png)
![SA-FedLoRA] Communic](https://aisholar.s3.ap-northeast-1.amazonaws.com/media/October2024/sa-fedlora-520x300.png)
![[SpliceBERT] A BERT](https://aisholar.s3.ap-northeast-1.amazonaws.com/media/July2024/splicebert-520x300.png)
![[IGModel] Methodolog](https://aisholar.s3.ap-northeast-1.amazonaws.com/media/July2024/igmodel-520x300.png)